European Open Science Cloud facilitates biodiversity research

Biodiversity data refers to all types of data utilized in ecology, genetics, and taxonomy. Most common data types include specimen data, nucleotide sequences, human observations, material samples, taxonomic backbones, and ecological data. Today, international workgroups have increasingly developed and managed large and sophisticated biodiversity datasets. Analyzing such a vast amount of data requires reliable biodiversity informatics tools.
It all started with fungi
PlutoF, an online platform for managing biodiversity data, was created by researchers at the University of Tartu, Estonia. It was designed to bridge the gap between the needs of contemporary research in biology and the existing software resources and databases. Originally, the PlutoF workbench was primarily meant for managing fungal biodiversity data. Today, however, this desktop is also equally used to collect data on animals, birds, plants, and insects.
“Fungal data are closely related to data from other groups of living organisms,” explains Kessy Abarenkov, Associate Professor in Biodiversity Informatics at the University of Tartu.
“For example, fungi can live in symbiosis with plants, cause disease in plants and animals, and so on. Therefore, it is natural that the information system must allow data storage on all groups of organisms, especially concerning the classification of living organisms.”
By making the system more universal, made it possible to successfully implement PlutoF to store and manage data from scientific collections and citizen science. PlutoF enables sharing datasets with any other user or workgroup. Thus, all projects can be connected to different data and datasets.
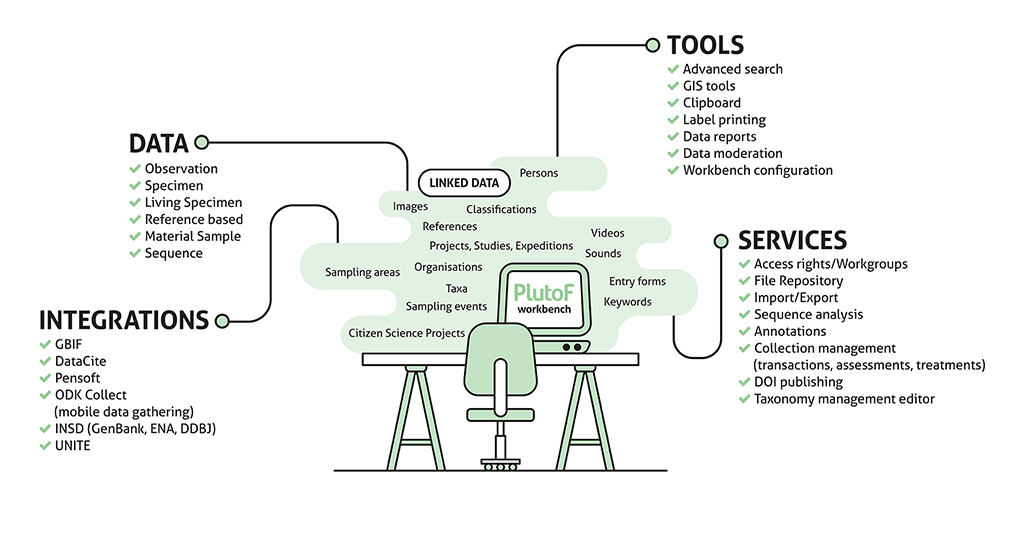
PlutoF workbench schema. Image: Kessy Abarenkov.
EOSC-Nordic will contribute to biodiversity research
“In the EOSC-Nordic project, we developed support for the use of Nordic computing centers and resources on the PlutoF workbench. This enables PlutoF users to run data analyses in high performance computing centers to which the users have access through their research projects. We also developed a tool to analyze fungal DNA sequences to compare particular data with similar data sets from researchers around the world. The EOSC-Nordic project has assisted potentially interested parties in testing our solutions. It has also helped to advertise better the e-services we offer,” says Kessy Abarenkov.
The collaboration network is growing
As of February 2022, PlutoF workbench has around 8,000 registered users from more than 100 countries. A few hundred users login to the system every day. It is clear that a large number of users will expand the collaboration network. For instance, a collaboration project of Estonian research infrastructure with the Estonian University of Life Sciences is currently underway. As part of this, the university will train data administrators to upload their research data to the PlutoF platform. Further data management is already taking place in PlutoF.
International cooperation is becoming more and more significant over time. “We have dozens of active partners. We work closely with marine researchers at the University of Gothenburg in the framework of the European ARMS programme, a network for genetic monitoring and early detection of non-indigenous marine species. The research collections of Latvian and Ukrainian universities have also started using PlutoF services, and this is a growing trend,” says Kessy Abarenkov.
Moreover, PlutoF is the basis for the web-based database and sequence management environment UNITE used for molecular identification of fungi.
Not just for researchers
A wide range of scientific areas can use biodiversity data to answer ecological and biogeographical questions, measure environmental impacts, identify threatened species and decide upon actions for their protection. The PlutoF platform currently manages data on more than 2,600 research projects. However, the system also enables any hobby nature observer to create a citizen science project to store and manage the data they collect. Observational data collected by hobby observers are available to researchers worldwide through the Global Biodiversity Information Facility (GBIF), an international network and data infrastructure providing open access to data about all types of life on Earth. Researchers often use data collected in the context of amateur science projects in their research. This practice is considered necessary.
The story behind the name
Kessy Abarenkov herself is very closely related to PlutoF, as the workbench development was part of her doctoral thesis. Interestingly, the choice of the workbench name coincided with a time when Pluto had just been removed from the list of planets, and the world press was actively covering the subject.
“As I began to feel sorry for the former planet, I decided to name the desktop after him. F was the first letter of the alphabet to match Pluto without infringing on Disney’s copyright,” Kessy Abarenkov explains.
Find PlutoF, the data management and publishing platform here.